TTK preliminary results
The TTK team is currently expanding the molecular library of redox active compounds beyond the pyridoxal database. The main aim is to build a structurally diverse set of molecules using the developed computational protocol, and apply machine learning techniques for high-throughput screening of the target 100 000 molecules.
The TTK team is primarily involved in work package WP1, which aims at developing a high-throughput screening methodology that enables the identification of promising candidates of water-soluble compounds for new generation redox flow batteries. The group has developed an efficient computational protocol that utilizes a combination of various electronic structure methods and it provides high quality predictions for reduction potentials. The protocol has been applied to build a molecular database comprising over 6700 pyridoxal-based molecules.
Several machine learning (ML) techniques, including the commonly used random forest algorithm as well as graph convolutional neural networks, were applied to the pyridoxal database to assess their performance for predicting reduction potentials and aqueous solubilities. Most of the tested ML methods were found to perform remarkably well, exceeding the accuracy of the quantum chemical computational protocol used to generate the pyridoxal database.
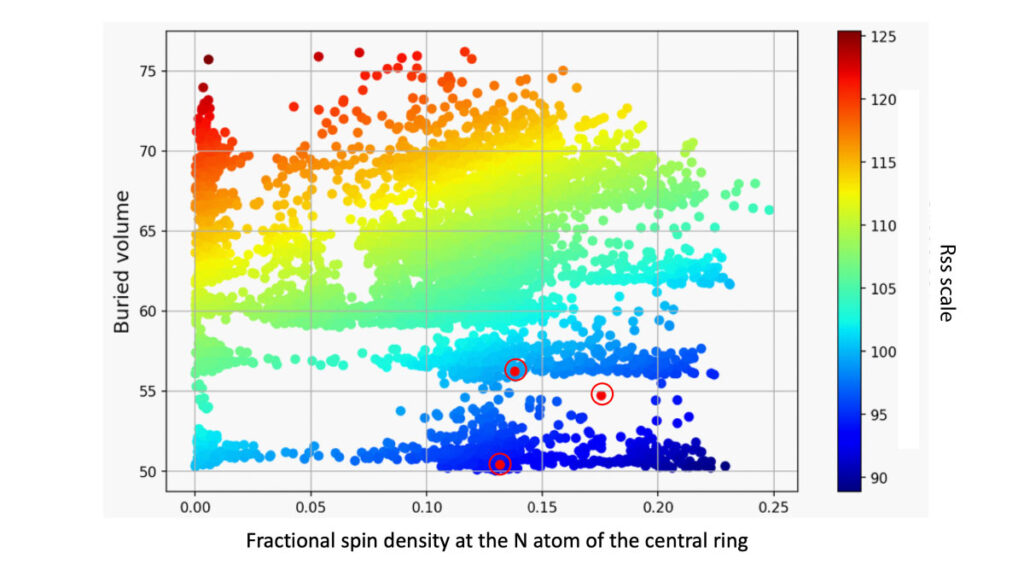
Some of the pyridoxal derivatives examined computational were synthesized by the JYU group, but stability problems were encountered in electrochemical studies. To provide insight into the stability issue, the TTK group adopted a new computational methodology for the characterization of radical stabilities, and provided a detailed analysis for the entire pyridoxal molecular set (see Figure below). The radical stability descriptors were found to be rather sensitive to the bulkiness of the substituent at the pyridinium N atom, as well as to electronic and steric nature of the other substituents. The radical forms of the compounds synthesized within work package WP4, were predicted to be unstable species.
Current activities of the TTK team are focused on the expansion of the molecular library of redox active compounds beyond the pyridoxal database. A molecular library of viologen derivatives is being developed, and a structurally very diverse set of molecules from open access databases are compiled. The application of the high-throughput screening methodology elaborated within CompBat will enable the screening of about 100 000 molecules.
Here we include a figure that illustrates the stability analysis of pyridoxal derivatives (see below).